|
Size: 6401
Comment:
|
Size: 11089
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
| = Scf = | = SCF = |
| Line 16: | Line 16: |
| Must input one of them if Hartree-Fock calculation is required. Require for restricted/unrestricted/restricted open shell Hartree-Fock calculations. | Must input one of them if Hartree-Fock calculation is required. Require for restricted/unrestricted/restricted open shell Hartree-Fock calculations. |
| Line 27: | Line 27: |
| Must input one of them if Kohn-Sham calculation is required. Require for restricted/unrestricted/restricted open shell Kohn-Sham calculations. | Must input one of them if Kohn-Sham calculation is required. Require for restricted/unrestricted/restricted open shell Kohn-Sham calculations. |
| Line 41: | Line 41: |
| Used in UHF\/ROHF\/UKS\/ROKS. Set number of alpha orbitals in each irreps. | Used in UHF\/ROHF\/UKS\/ROKS. Set number of beta orbitals in each irreps. |
| Line 59: | Line 59: |
Commonly used functionals: SVWN5, BLYP, B3LYP, CAM-B3LYP, etc. }}} {{{#!wiki LSDA: ex_fun=1 co_fun=1 SVWN5: ex_fun=1 co_fun=2 PW91: ex_fun=3 co_fun=3 SAOP: ex_fun=30 co_fun=30 BLYP: ex_fun=4 co_fun=8 BHHLYP ex_fun=21 co_fun=8 B2PLYP ex_fun=22 co_fun=8 B3LYP ex_fun=20 co_fun=0 LC-BLYP ex_fun=104 co_fun=8 CAM-B3LYP alpha=0.19d0 beta=0.46d0 ex_fun=120 co_fun=0 B3PW91 ex_fun=27 co_fun=0 PBE PBE0 VBLYP GBLYP SF5050 LC-BVWN5 LC-BLYP SAOP |
|
| Line 90: | Line 140: |
| None Direct SCF, do not use direct grid. | None Direct SCF, do not use direct grid. |
| Line 102: | Line 152: |
| Line 104: | Line 155: |
| If the numerical integral $\rho_{\mu}<threshRho$, the basis $\chi_{\mu}$ will be neglected. The $\rho_{\mu}$ is defined as \begin{align*} \rho_{\mu}=\sum_i w_i*\chi_{\mu}(r_i)\chi_{\mu}(r_i) \end{align*} Default value: $ThreshRho=\frac{thresh\_ene}{maxradgrid*maxanggrid*natom}$ }}} == SCF convergence === === Maxiter === {{{#!wiki Number of maxim iterations in SCF. }}} === Nodiis === {{{#!wiki Disable DIIS. }}} |
If the numerical integral <<latex($\rho_{\mu}<threshRho$)>>, the basis <<latex($\chi_{\mu}$)>> will be neglected. The <<latex($\rho_{\mu}$)>> is defined as <<latex($\rho_{\mu}=\sum_i w_i*\chi_{\mu}(r_i)\chi_{\mu}(r_i)$)>> Default value: <<latex($ThreshRho=\frac{thresh\_{ene}}{maxradgrid*maxanggrid*natom}$)>> }}} === Coulpot/Coulpotlmax/Coulpottol === {{{#!wiki Use multipolar expansion to get coulomb potential and matrix. }}} === Coulpotlmax === {{{#!wiki Max L value for coulomb potential multipolar expansion. Default value: 5 }}} === Coulpottol === {{{#!wiki Cutoff threshold parameter for coulomb potential multipolar expansion, more higher more accurate. Default value: 8. }}} == SCF convergence == === MAXITER === {{{ The maximum Number of SCF iteration }}} === NODIIS === {{{ Logical control parameter. Disable DIIS. }}} |
| Line 127: | Line 191: |
| Line 132: | Line 195: |
| Line 137: | Line 199: |
| Line 140: | Line 201: |
| Convergence threhhold. | Convergence threhhold. |
| Line 143: | Line 204: |
| Line 148: | Line 208: |
| === DIISmode === {{{ DIISmode: 0: diisdim goes from 0 to maxdiis, then cycles to 0. And reset to 0 when diis fails. 1: diisdim goes from 0 to maxdiis, keeps maxdiis. And throw the oldest vector (reduce diisdim) when diis fails. Default: 0. }}} |
|
| Line 153: | Line 219: |
| Line 158: | Line 223: |
| Line 163: | Line 227: |
=== iAufbau === {{{#!wiki Enable or disable Aufbau law to change orbital occupation number in SCF iteration. }}} FOA FCA FVA |
=== IAUFBAU === {{{ Control parameter of electron occupation protocol in each SCF iteration. IAUFBAU = 1, electron occupation obeys Aufbau principle(default); IAUFBAU = 2, electrons complies with specific occupation pattern based on maximum occupation method(mom); IAUFBAU = 3, electrons complies with specific occupation pattern based on maximum occupation method(mom).Update MO coefficients and reorder occupied orbitals in each iteration. WARNING if IAUFBAU=2 or 3 without initial guess=read (this means initial guess is bad), the result is unpredictable. }}} == Print and output SCF orbital into Molden format == === print === {{{#!wiki Print level. }}} === iprtmo === {{{#!wiki Require to print MO coefficients. Values: 1 Only print orbital energy and occupation numbers. 2 Print all information. }}} === Molden === {{{#!wiki Output SCF orbital into Molden format file. }}} |
| Line 179: | Line 257: |
| Line 183: | Line 260: |
| delta P will be used in integral prescreening instead of P. | delta P will be used in integral prescreening instead of P. |
| Line 186: | Line 263: |
| Line 191: | Line 265: |
| For debugging. Set a stick threshhold for integral prescreening directly. Thresh\_ene*1.d-4. }}} === Nok2Prim === |
For debugging. Set a strict threshold (thresh_rho=1.d-4) for integral prescreening directly. }}} === Nok2Prim === |
| Line 197: | Line 270: |
| value and perform screening. | value and perform screening. |
| Line 201: | Line 274: |
| Line 211: | Line 283: |
| Line 216: | Line 287: |
| Line 221: | Line 291: |
| Line 226: | Line 295: |
| If Read is used, the old orbital will be read and there is an extra line to set files contain old orbital. The filename is INPORB. It is generated by previous SCF calcualtion with the name Task.scforb }}} === print === {{{#!wiki Print level. }}} === iprtmo === {{{#!wiki Require to print MO coefficients. Values: 1 Only print orbital energy and occupation numbers. 2 Print all information. }}} |
If Read is used, the old orbital will be read. The old orbital saves in a file named "inporb" in BDF_TMPDIR . It is generated by previous SCF calcualtion with the name Task.scforb. }}} |
| Line 246: | Line 301: |
| Values: -1 Do not cut tail. 1 Project a LMO into fragment with largest Lowdin population. | Values: -1 Do not cut tail. 1 Project a LMO into fragment with largest Lowdin population. |
| Line 254: | Line 309: |
| === Keyword4 === {{{#!wiki xxx |
=== CHECKLIN === {{{#!wiki Check if the basis sets is linear dependent. If diffuse basis set is used, SCF do not converge or ridiculous energy observed, it is better to check linear dependent of the basis set. }}} === ΤΟLLIN === {{{#!wiki Tolerance of basis set linear dependent. Default value 1.d-7. }}} === ifPair === {{{#!wiki used to excite electrons (MOM)with following keywords:<<BR>> hpalpha,hpbeta<<BR>> then with number of partical-hole pairs N<<BR>> then with 2N lines specificate partical-hole pairs. (0 is do nothing, indexes start from 1)<<BR>> eg. the molecular is has 4 irreducible representation, we want to excite electrons from orb 5,6 to 8,9 in rep 1 and 3 to 4 in rep 3 (alpha) & 7 to 8 in rep 1(beta): }}} {{{ ifpair hpalpha 2 5 0 3 0 8 0 4 0 6 0 0 0 9 0 0 0 hpbeta 1 7 0 0 0 8 0 0 0 }}} {{{#!wiki this should be combined with iaufbau=2 or 3.<<BR>> WARNING: this function will not check whether partical orbital is filled or whether hole orbiltal is not filled. }}} === pinalpha , pinbeta === {{{#!wiki specificate fix orbitals<<BR>> first line specificates the number of fix orbitals<<BR>> then with N lines specificate fix orbitals. (0 is do nothing, indexes start from 1)<<BR>> (somewhat likes hpalpha/hpbeta input)<<BR>> these keywords leads to SCF_solver from solve FC=SCE to <<latex($\tilde{F}U=UE,\tilde{F}=C^\dagger FC$)>> <<BR>> |
| Line 264: | Line 358: |
| == How to perform a direct DFT calculation with B3LYP functional? == {{{ $COMPASS Title Cocaine Molecule test run, CC-PVDZ Basis CC-PVDZ Geometry XYZ # The molecule geometry will be read from file $BDFTASK.xyz End Geometry Skeleton # This keyword must be used. $End $xuanyuan Direct # Direct SCF. Schwarz # Schwarz prescreening. $end $scf RKS DFT functional B3LYP Molden # This keyword is used to output SCF orbital to molden format file. $end }}} == How to read molecular orbital as initial guess orbital or restart SCF calculation? == Suppose you have performed a calculation and generated aSCF orbital file in your work directory as test.scforb. Usually, this file atomically generated by SCF module. This file also can be used to restart SCF calculation via read it as initial guess orbital. {{{ $COMPASS Title Cocaine Molecule test run, CC-PVDZ Basis CC-PVDZ Geometry XYZ # The molecule geometry will be read from file $BDFTASK.xyz. End Geometry Skeleton # This keyword must be used. $End $xuanyuan Direct # Direct SCF. Schwarz # Schwarz prescreening. $end # Copy orbital file test.scforb as inporb in BDF_TMPDIR % cp $BDF_WORKDIR/test.scforb $BDF_TMPDIR/inporb $scf RKS DFT functional B3LYP Guess # Read orbital as initial guess orbital Read Molden # This keyword is used to output SCF orbital to molden format file. $end }}} |
SCF
Contents
HF/DFT.
General keywords
RHF/UHF/ROHF
- Must input one of them if Hartree-Fock calculation is required. Require for restricted/unrestricted/restricted open shell Hartree-Fock calculations.
Example:
$Scf RHF $end
RKS/UKS/ROKS
- Must input one of them if Kohn-Sham calculation is required. Require for restricted/unrestricted/restricted open shell Kohn-Sham calculations.
Occupy
- Used in RHF/RKS. Set double occupied number of each irreps. The following line is an integer array, $noccu(i),i=1,\cdots, nirreps$.
Alpha
- Used in UHF/ROHF/UKS/ROKS. Set number of alpha orbitals in each irreps. The following line is an integer array, $nalpha(i),i=1,\cdots, nirreps$.
Beta
- Used in UHF\/ROHF\/UKS\/ROKS. Set number of beta orbitals in each irreps. The following line is an integer array,$ nbeta(i),i=1,\cdots , nirreps$.
Charge
- Charge of the state.
Spin
- Spin of the state. The value is 2S+1.
keyword
DFT functional keywords
DFT
- DFT functional used in Korn-Sham calculation. Commonly used functionals: SVWN5, BLYP, B3LYP, CAM-B3LYP, etc.
- LSDA:
- ex_fun=1 co_fun=1
- ex_fun=1 co_fun=2
- ex_fun=3 co_fun=3
- ex_fun=30 co_fun=30
- ex_fun=4 co_fun=8
- ex_fun=21 co_fun=8
- ex_fun=22 co_fun=8
- ex_fun=20 co_fun=0
- ex_fun=104 co_fun=8
- alpha=0.19d0 beta=0.46d0 ex_fun=120 co_fun=0
- ex_fun=27 co_fun=0
RS
- Alpha and beta value in CAM calculation. The following line are two float number. For example : 0.33 0.15
D3
- Grimmers dispersion corrrection for DFT.
DFT grid keywords
NPTRAD
- Number of radius grid points.
NPTANG
- Number of angular grid points.
Grid
- Set DFT grid. Support values are: Ultra Coarse, Coarse, Medium, Fine, Ultra Fine, SG1.
NoSymGrid
- Do not use symmetry dependent grid. Only for debugging.
DirectGrid
Use DirectGrid. Basis set values on the grid points are calculated directly. Default: Direct SCF, use direct grid. None Direct SCF, do not use direct grid.
NoDirectGrid
- Force to do not use direct grid.
NoGridSwitch
- For direct SCF, DFT grid can be switched. At the beging of iteration, Ultra coase grid will be used. After energer change is little than a value, such as 1.d-4, the medium grid or user setted grid
will be used. NoGridSwitch dissiable grid switch and use default grid directly.
ThreshRho
If the numerical integral
 , the basis
, the basis 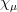 will be neglected. The
will be neglected. The  is defined as
is defined as Default value:

Coulpot/Coulpotlmax/Coulpottol
- Use multipolar expansion to get coulomb potential and matrix.
Coulpotlmax
- Max L value for coulomb potential multipolar expansion. Default value: 5
Coulpottol
- Cutoff threshold parameter for coulomb potential multipolar expansion, more higher more accurate. Default value: 8.
SCF convergence
MAXITER
The maximum Number of SCF iteration
NODIIS
Logical control parameter. Disable DIIS.
MaxDiis
- Maxim number of Diis space. Default: 8
THRENE
- Convergence threshhold for energy. Default: 1.d-8.
THRDEN
- Convergence threshhold for density matrix. Default: 3.d-6.
ThreshConverg
- Convergence threhhold. Two float value: DeltaE DeltaD
THRDIIS
- Threshold to turn on DIIS. Default: 0.15.
DIISmode
DIISmode: 0: diisdim goes from 0 to maxdiis, then cycles to 0. And reset to 0 when diis fails. 1: diisdim goes from 0 to maxdiis, keeps maxdiis. And throw the oldest vector (reduce diisdim) when diis fails. Default: 0.
Vshift
- Level shift value.
Damp
- Damping value.
Icheck
- Check Aufbau law.
IAUFBAU
Control parameter of electron occupation protocol in each SCF iteration. IAUFBAU = 1, electron occupation obeys Aufbau principle(default); IAUFBAU = 2, electrons complies with specific occupation pattern based on maximum occupation method(mom); IAUFBAU = 3, electrons complies with specific occupation pattern based on maximum occupation method(mom).Update MO coefficients and reorder occupied orbitals in each iteration. WARNING if IAUFBAU=2 or 3 without initial guess=read (this means initial guess is bad), the result is unpredictable.
Print and output SCF orbital into Molden format
- Print level.
iprtmo
- Require to print MO coefficients. Values: 1 Only print orbital energy and occupation numbers. 2 Print all information.
Molden
- Output SCF orbital into Molden format file.
Expert keywords
IfNoDeltaP
- Dissable using DeltaP to update Fock matrix.
IfDeltaP
- Delta P is used to update density matrix. In direct SCF calculation, delta P will be used in integral prescreening instead of P. Default: true.
Optscreen
- For debugging. Set a strict threshold (thresh_rho=1.d-4) for integral prescreening directly.
Nok2Prim
- Disable primative integral screenning via K2 integrals. Use (SS|SS) esitimating primative integral value and perform screening. Default: Direct SCF, use K2 primative screening.
- None Direct SCF, use (SS|SS) integral.
FixDif
- Fix factor for incremental fock update. If the factor is not fixed, use the formular \begin{align*}
fac=1-\frac{D{n+1}-Dn}{D{n+1}*D{n+1}} \\ F{n+1}=Fn+fac*\delta F
Jengin
- Use Jengin method calculate J matrix. In debugging, not support now.
LinK
- Use LinK calculate K matrix. In debugging, not support now.
Guess
- Method to get initial guess orbital. The following line is a string.
- Values: Atom, Hcore, Huckel, Read. If Read is used, the old orbital will be read. The old orbital saves in a file named "inporb" in BDF_TMPDIR . It is generated by previous SCF calcualtion with the name Task.scforb.
Cutlmotail
- Methods to cut long Coulomb tails of Local molecular orital. Values: -1 Do not cut tail. 1 Project a LMO into fragment with largest Lowdin population.
- 2 Similar with 1, but project a LMO into predefined group of fragments with largest Lowdin population. 3 Very stick cutoff. Project LMO to a fragment plus several atoms. The threshhold is 1.d-4.
- SCF calcualtion based on LMO because diffirent fragment interaction policy can be predefined, which will reduce ERIs need to be calculated.
CHECKLIN
Check if the basis sets is linear dependent. If diffuse basis set is used, SCF do not converge or ridiculous energy observed, it is better to check linear dependent of the basis set.
ΤΟLLIN
Tolerance of basis set linear dependent. Default value 1.d-7.
ifPair
used to excite electrons (MOM)with following keywords:
hpalpha,hpbeta
then with number of partical-hole pairs N
then with 2N lines specificate partical-hole pairs. (0 is do nothing, indexes start from 1)
eg. the molecular is has 4 irreducible representation, we want to excite electrons from orb 5,6 to 8,9 in rep 1 and 3 to 4 in rep 3 (alpha) & 7 to 8 in rep 1(beta):
ifpair hpalpha 2 5 0 3 0 8 0 4 0 6 0 0 0 9 0 0 0 hpbeta 1 7 0 0 0 8 0 0 0
this should be combined with iaufbau=2 or 3.
WARNING: this function will not check whether partical orbital is filled or whether hole orbiltal is not filled.
pinalpha , pinbeta
specificate fix orbitals
first line specificates the number of fix orbitals
then with N lines specificate fix orbitals. (0 is do nothing, indexes start from 1)
(somewhat likes hpalpha/hpbeta input)
these keywords leads to SCF_solver from solve FC=SCE to 
Depend Files
Filename |
Description |
Format |
|
|
|
Examples
How to perform a direct DFT calculation with B3LYP functional?
$COMPASS Title Cocaine Molecule test run, CC-PVDZ Basis CC-PVDZ Geometry XYZ # The molecule geometry will be read from file $BDFTASK.xyz End Geometry Skeleton # This keyword must be used. $End $xuanyuan Direct # Direct SCF. Schwarz # Schwarz prescreening. $end $scf RKS DFT functional B3LYP Molden # This keyword is used to output SCF orbital to molden format file. $end
How to read molecular orbital as initial guess orbital or restart SCF calculation?
Suppose you have performed a calculation and generated aSCF orbital file in your work directory as test.scforb. Usually, this file atomically generated by SCF module. This file also can be used to restart SCF calculation via read it as initial guess orbital.
$COMPASS Title Cocaine Molecule test run, CC-PVDZ Basis CC-PVDZ Geometry XYZ # The molecule geometry will be read from file $BDFTASK.xyz. End Geometry Skeleton # This keyword must be used. $End $xuanyuan Direct # Direct SCF. Schwarz # Schwarz prescreening. $end # Copy orbital file test.scforb as inporb in BDF_TMPDIR % cp $BDF_WORKDIR/test.scforb $BDF_TMPDIR/inporb $scf RKS DFT functional B3LYP Guess # Read orbital as initial guess orbital Read Molden # This keyword is used to output SCF orbital to molden format file. $end
